
4. Algorithm for visualising the model overlaid on high-dimensional data
Source:vignettes/quollr4algo.Rmd
quollr4algo.RmdThis walks through the algorithm for constructing a model from 2-D embedding data and visualising it alongside high-dimensional data. The process involves two major steps:
- Constructing the model in the 2-D embedding space using hexagonal binning and triangulation.
- Lifting the model into high dimensions to link it back to the original data space.
Step 1: Construct the 2-D model
Hexagonal Binning
To begin, we preprocess the 2-D embedding and create hexagonal bins over the layout.
## To pre-process the data
nldr_obj <- gen_scaled_data(nldr_data = scurve_umap)
## Obtain the hexbin object
hb_obj <- hex_binning(
nldr_scaled_obj = scurve_model_obj$nldr_scaled_obj,
b1 = 21, q = 0.1)
all_centroids_df <- hb_obj$centroids
counts_df <- hb_obj$std_ctsExtract bin centroids
Next, we extract the centroid coordinates and standardised bin counts. These will be used to identify densely populated regions in the 2-D space.
## To extract all bin centroids with bin counts
df_bin_centroids <- merge_hexbin_centroids(centroids_data = all_centroids_df,
counts_data = counts_df)
benchmark_highdens <- 0
## To extract high-densed bins
model_2d <- df_bin_centroids |>
dplyr::filter(n_h > benchmark_highdens)
glimpse(model_2d)
#> Rows: 251
#> Columns: 5
#> $ h <int> 58, 68, 69, 70, 71, 72, 73, 78, 79, 90, 91, 92, 93, 94, 95, 96, 98…
#> $ c_x <dbl> 0.7804473, 0.1641342, 0.2228307, 0.2815272, 0.3402237, 0.3989202, …
#> $ c_y <dbl> -0.01401484, 0.03681781, 0.03681781, 0.03681781, 0.03681781, 0.036…
#> $ n_h <dbl> 4, 1, 5, 6, 9, 5, 3, 3, 6, 7, 5, 2, 1, 5, 9, 1, 1, 5, 6, 10, 1, 4,…
#> $ w_h <dbl> 0.004, 0.001, 0.005, 0.006, 0.009, 0.005, 0.003, 0.003, 0.006, 0.0…Triangulate the bin centroids
We then triangulate the hexagon centroids to build a wireframe of neighborhood relationships.
## Wireframe
tr_object <- tri_bin_centroids(centroids_data = df_bin_centroids)
str(tr_object)
#> List of 2
#> $ trimesh_object:List of 11
#> ..$ n : int 588
#> ..$ x : num [1:588] -0.1 -0.0413 0.0174 0.0761 0.1348 ...
#> ..$ y : num [1:588] -0.116 -0.116 -0.116 -0.116 -0.116 ...
#> ..$ nt : int 1106
#> ..$ trlist: int [1:1106, 1:9] 1 23 43 22 22 3 45 23 23 64 ...
#> .. ..- attr(*, "dimnames")=List of 2
#> .. .. ..$ : NULL
#> .. .. ..$ : chr [1:9] "i1" "i2" "i3" "j1" ...
#> ..$ cclist: num [1:1106, 1:5] -0.0707 -0.0413 -0.1293 -0.0707 -0.0413 ...
#> .. ..- attr(*, "dimnames")=List of 2
#> .. .. ..$ : NULL
#> .. .. ..$ : chr [1:5] "x" "y" "r" "area" ...
#> ..$ nchull: int 68
#> ..$ chull : int [1:68] 1 2 3 4 5 6 7 8 9 10 ...
#> ..$ narcs : int 1693
#> ..$ arcs : int [1:1693, 1:2] 2 22 1 2 23 22 43 44 22 23 ...
#> .. ..- attr(*, "dimnames")=List of 2
#> .. .. ..$ : NULL
#> .. .. ..$ : chr [1:2] "from" "to"
#> ..$ call : language tri.mesh(x = centroids_data[["c_x"]], y = centroids_data[["c_y"]])
#> ..- attr(*, "class")= chr "triSht"
#> $ n_h : num [1:588] 0 0 0 0 0 0 0 0 0 0 ...Generate edges from triangulation
Using the triangulation object, we generate edges between centroids. We retain only edges connecting densely populated bins.
trimesh_data <- gen_edges(tri_object = tr_object, a1 = hb_obj$a1) |>
dplyr::filter(from_count > benchmark_highdens,
to_count > benchmark_highdens)
## Update the edge indexes to start from 1
trimesh_data <- update_trimesh_index(trimesh_data)
glimpse(trimesh_data)
#> Rows: 655
#> Columns: 10
#> $ from <int> 69, 68, 70, 69, 90, 90, 110, 131, 131, 111, 91, 91, 111…
#> $ to <int> 90, 69, 91, 70, 111, 91, 132, 152, 132, 112, 92, 112, 1…
#> $ x_from <dbl> 0.2228307, 0.1641342, 0.2815272, 0.2228307, 0.1934824, …
#> $ y_from <dbl> 0.03681781, 0.03681781, 0.03681781, 0.03681781, 0.08765…
#> $ x_to <dbl> 0.1934824, 0.2228307, 0.2521789, 0.2815272, 0.2228307, …
#> $ y_to <dbl> 0.08765046, 0.03681781, 0.08765046, 0.03681781, 0.13848…
#> $ from_count <dbl> 5, 1, 6, 5, 7, 7, 4, 1, 1, 6, 5, 5, 6, 7, 6, 6, 3, 3, 5…
#> $ to_count <dbl> 7, 5, 5, 6, 6, 5, 7, 3, 7, 6, 2, 6, 3, 3, 1, 9, 7, 3, 1…
#> $ from_reindexed <int> 3, 2, 4, 3, 10, 10, 22, 35, 35, 23, 11, 11, 23, 36, 24,…
#> $ to_reindexed <int> 10, 3, 11, 4, 23, 11, 36, 49, 36, 24, 12, 24, 37, 50, 2…Visualise the triangular mesh
trimesh <- ggplot(model_2d, aes(x = c_x, y = c_y)) +
geom_trimesh() +
coord_equal() +
xlab(expression(C[x]^{(2)})) + ylab(expression(C[y]^{(2)})) +
theme(axis.text = element_text(size = 5),
axis.title = element_text(size = 7))
trimesh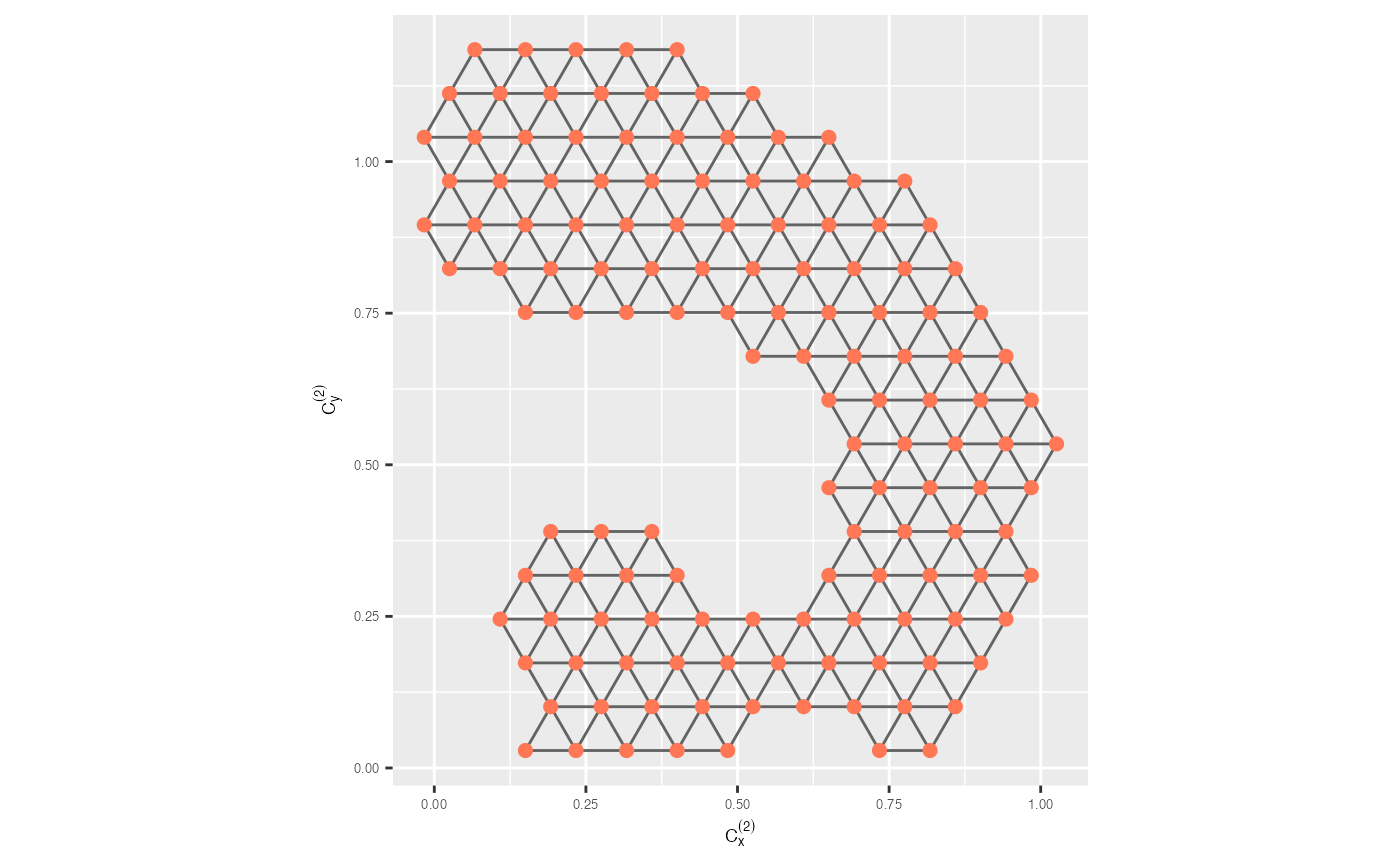
Step 2: Lift the model into high dimensions
Map bins to high-dimensional observations
We begin by extracting the original data with their assigned hexagonal bin IDs.
nldr_df_with_hex_id <- hb_obj$data_hb_id
glimpse(nldr_df_with_hex_id)
#> Rows: 1,000
#> Columns: 4
#> $ emb1 <dbl> 0.27708147, 0.69717161, 0.77934921, 0.17323121, 0.21793445, 0.593…
#> $ emb2 <dbl> 0.91343544, 0.53767948, 0.39861033, 0.95285002, 0.98320848, 1.048…
#> $ ID <int> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19…
#> $ h <int> 427, 287, 226, 446, 468, 495, 132, 292, 415, 229, 289, 91, 155, 4…Compute high-dimensional coordinates for bins
We calculate the average high-dimensional coordinates for each bin and retain only the ones matching the 2-D model bins.
model_highd <- avg_highd_data(highd_data = scurve, scaled_nldr_hexid = nldr_df_with_hex_id)
model_highd <- model_highd |>
dplyr::filter(h %in% model_2d$h)
glimpse(model_highd)
#> Rows: 251
#> Columns: 8
#> $ h <int> 58, 68, 69, 70, 71, 72, 73, 78, 79, 90, 91, 92, 93, 94, 95, 96, 98,…
#> $ x1 <dbl> -0.37105188, 0.95812568, 0.85450337, 0.73126005, 0.47416208, 0.2648…
#> $ x2 <dbl> 1.90644261, 0.08539250, 0.09171102, 0.12907776, 0.10794141, 0.12362…
#> $ x3 <dbl> 1.922623, 1.286348, 1.507735, 1.676232, 1.877526, 1.962058, 1.99642…
#> $ x4 <dbl> -0.0082718681, 0.0026502232, 0.0051225053, -0.0043335180, -0.002598…
#> $ x5 <dbl> 0.0018876029, 0.0170739819, 0.0003245598, 0.0021079776, 0.000127695…
#> $ x6 <dbl> 0.016977853, 0.087550004, -0.013016748, -0.035630709, 0.007849772, …
#> $ x7 <dbl> 0.0028050043, -0.0024864689, -0.0039496434, -0.0024049571, 0.001697…Step 3: Visualise the high-dimensional model
We now combine all components—high-dimensional data, the 2-D model, lifted high-dimensional centroids, and the triangulation—and render the model using an interactive tour.
Prepare data for visualisation
df_exe <- comb_data_model(highd_data = scurve,
model_highd = model_highd,
model_2d = model_2d)Interactive tour of model overlay
tour1 <- show_langevitour(point_data = df_exe, edge_data = trimesh_data)
tour1